Two weeks before Christmas, BBC reported of a new SARS-Cov-2 variant spreading in Southeastern England1. Termed lineage B117 or VUI-202012/01, this SARS-Cov-2 variant was first identified in Kent County, England on September 20th and reached 1108 cases as of December 13th, 20202. In response to the strain’s emergence, Canada became one of several countries to halt all flights to the UK for 72 hours to minimize the risk of international spread3. Despite these measures to prevent the UK strain from coming to Canada, 14 of the SARS-Cov-2 cases in Canada now are positive for the British variant4.
A SARS-Cov-2 variant is a subtype of SARS-Cov-2 that’s genetically distinct. These variants emerge as a result of genetic mutations. Microbes get gene mutations all the time. In fact, 5775 (and counting) distinct genome variants SARS-Cov-2 have been detected since the COVID-19 pandemic began4. Before I can talk about the mutations in more detail, I need to define what proteins are. Most genes encode for a single protein. In biology, a protein isn’t the meat that you eat during a meal. Instead, think of a protein as a machine that has a specific function. These proteins can help turn the food we eat into nutrients, keep us safe from disease, and keep all our body parts functioning. In the same way, the genes within SARS-Cov-2 encode for proteins that enable the virus to attack us and cause disease.
Given how there’s so many SARS-Cov-2 genetic variants, how have researchers classified the UK variant that’s come to Canada? Just before the Christmas break, my PhD supervisor shared a link to a virology forum post authored by the members of the COVID-19 Genomics UK Consortium5. If you follow the link and see Figure 1, you’ll notice that there’s a group of circles and lines placed within a large, grey rectangle. What you’re seeing in that figure is a phylogenetic tree. Each circle in this tree represents an individual SARS-Cov-2 isolate obtained from a patient. The lines connecting the circles represent branches that tell us how similar the isolates are with each other based on their genetic material. The length of the branches also tells us how far apart the isolates are from each other based on how much time the SARS-Cov-2 isolates had to mutate on their own. If the isolates are located in similar branches, or clusters, they are more closely related. The long length of the branch connecting the B117 lineage shows that these isolates mutated a lot compared to the other isolates in the UK. It’s this kind of branch that tells us that there’s a new set of SARS-Cov-2 variants that’s concentrated in southeastern England.
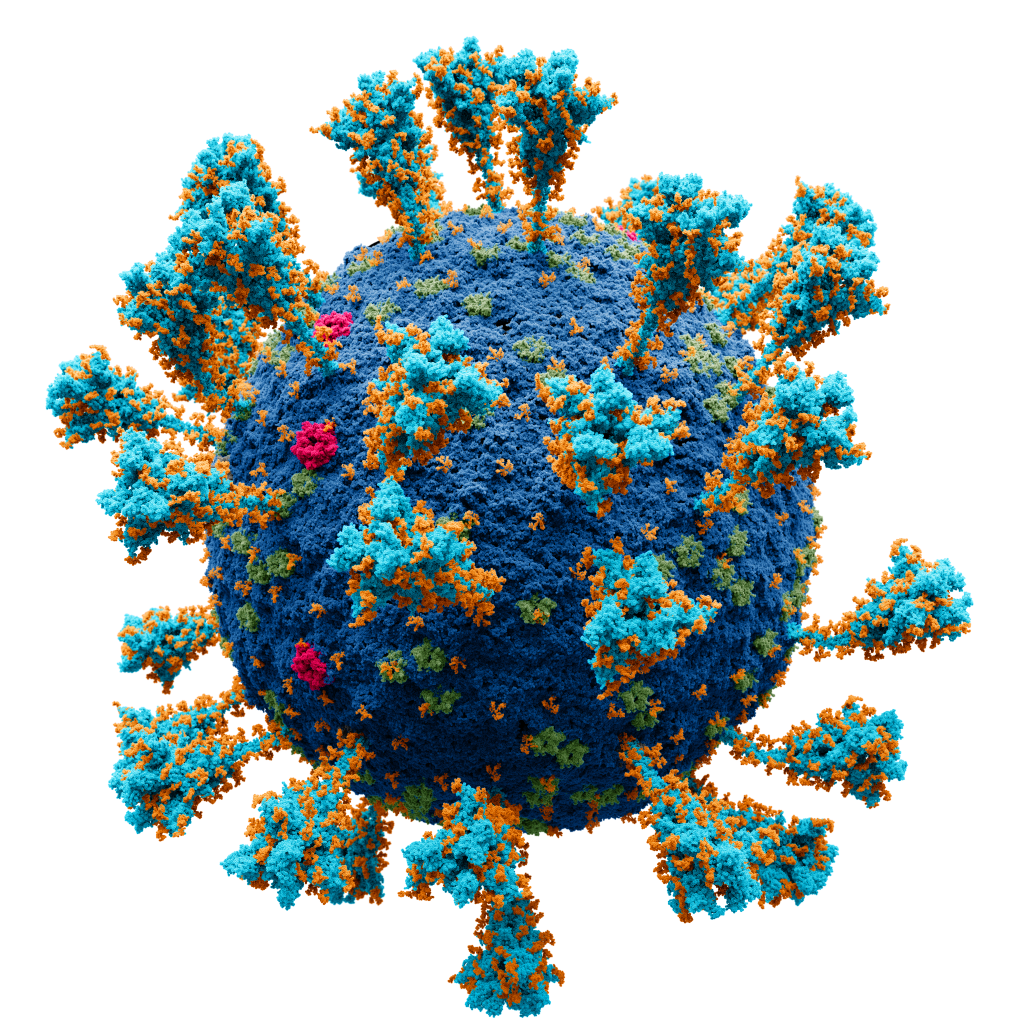
Now, every tree also has roots. If you look at Figure 2, you’ll see that the consortium also measured how much the B117 lineage’s genomes diverged from the root of the tree relative to the rest of isolates. While the rates of gene sequence evolution are similar between the two sets of isolates, it’s clear that the B117 class of variants did mutate faster than the non-B117 variants. The box that the arrow points to from the long branch indicates that many of these mutations are taking place at genes associated with the spike protein (the red shapes on the SARS-Cov-2 figure). The spike protein is the protein that the virus uses to bind and enter airway cells to drive respiratory infection.
Most mutations are deleterious, meaning that they provide negative effects on the microbe’s ability to proliferate in an environment. However, mutations that confer benefits for the microbe do exist. Such is the case with the B117 SARS-Cov-2 variant. Two of these mutations, N501Y and P681H, were observed in the isolates of the B117 variant. Both of these mutations help SARS-Cov-2 enter lung cells more easily and make it easier for other people to get COVID-19. The post goes on to note that these two mutations have never been observed together in the same variants until now. The presence of these mutations may explain why this new variant was observed to be 56% more transmissible relative to other SARS-Cov-2 variants6.
There are still many questions concerning the new SARS-Cov-2 variant from the UK. How did the UK variant mutate so quickly? Will patients infected with this new variant experience more severe COVID-19 symptoms? While initial reports suggest that it’s extremely unlikely the new variant will defeat current vaccines7, how likely and frequently will we see SARS-Cov-2 mutants that do beat current vaccines? The more we know about something, the more questions we ask.
Such questions represent the frustrations of learning about something that mutates so quickly. But I also want to see the opportunities behind these questions. The current pandemic has given us a wake up call to learn more about viruses and other infective agents. Answering these questions will go a long way towards an optimized response to the current pandemic and future infectious disease outbreaks.
- BBC News. December 15th, 2020. https://www.bbc.com/news/health-553082116.
- The UK Government. December 14th, 2020. https://www.gov.uk/government/news/phe-investigating-a-novel-strain-of-covid-19
- The Globe and Mail. December 20th, 2020. https://www.theglobeandmail.com/world/article-canada-halts-flights-from-uk-as-fast-spreading-variants-of-covid-19/
- The Globe and Mail. January 12th, 2021 https://www.theglobeandmail.com/canada/article-ontario-issues-stay-at-home-order-as-covid-cases-rise/
- Koyama, T., Platt, D., and Parida, L. (2020). Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ 98, 495–504.
- (2020a). Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563
- (2020b). Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. https://cmmid.github.io/topics/covid19/uk-novel-variant.html
- The Guardian. January 10th, 2021 https://www.theguardian.com/science/2021/jan/10/uk-covid-variant-unlikely-to-evade-vaccines
I had this post about the UK variant prepared since December, but given the rapidly changing nature of the COVID-19 pandemic, I wanted to wait until we had some preliminary information about the UK variant. Given that the UK variant has come to Canada, I thought that now was the best time to share this blog post.
I still have special interviews coming up, but I’m waiting to get the responses to the interview questions to put into my blog. In the meantime, I hope this post helped you gain some insights into the UK variant that’s come to Canada.
Finally, you may be noticing a lot of SARS-Cov-2 content on my blog as of late. This was done to keep up with the latest news on SARS-Cov-2 and explain it to the community. I have non-COVID-19 posts coming up, but I’d like to release those once I figure out the interviews.