Omicron BA.2 – a descendent of the original Omicron
Just as Omicron infection counts are starting to decline in Canada, the first COVID-19 cases featuring the Omicron sublineage BA.2 – which has been detected in 40 countries already – have been found in Canada1. As of last night, 51 cases of Omicron BA.2 have been detected in Canada2, and 426 in the UK3. I previously summarized the evolutionary history of the Omicron variant as documented by the South African lab that first discovered Omicron4. In that study, the researchers identified and sequenced three Omicron sublineages: BA.1, BA.2, and BA.3. The BA.2 variant that’s topping news headlines right now is the same BA.2 variant I talked about in that blog post. With that in mind, BA.2 has likely been present in multiple countries as early as November 2021.
BA.2 is a descendent lineage from BA.1, meaning that BA.2 shares some mutations observed in BA.1. However, BA.2 has other mutations that make this sublineage unique. Most notably, the amino acids that were deleted at positions 69 and 70 in BA.1’s spike protein reappear in BA.23. One major concern with SARS-Cov-2 monitoring now is that the TaqPath COVID-19 test kit5 used to detect Omicron relies on identifying the deletion at amino acids 69 and 70 in the spike protein. The reemergence of these amino acids makes it harder for scientists to know which Omicron sublineage is infecting a person with the polymerase chain reaction (PCR) alone. To be clear, PCR assays are still more than capable of detecting SARS-Cov-2 by detecting the genes found in all SARS-Cov-2 genomes. Nonetheless, sequencing the whole genome becomes necessary when distinguishing between the BA.1 and BA.2 lineages as of today. It’s because of these traits that BA.2 has been dubbed “Stealth Omicron”. However, the fact that the scientific community already knew about BA.2 as far back as November 2021 makes this term a misnomer in my opinion.
As wide reporting on the BA.2 sublineage in Canada and the rest of the world has recently begun, it is difficult to know for certain how BA.2 will affect current infection rates and hospitalization counts. The first clues of how BA.2 infection could affect COVID-19 cases and outcomes can be found in Denmark. Anders Fomsgaard, a virologist at the State Serum Institute in Denmark, reported that BA.2 has dominated Omicron-based infections in Denmark and that hospitalizations continue to decrease despite BA.2 being the dominant Omicron sublineage6. As for how well vaccines work against BA.2, more time is needed to know whether a difference in vaccine efficacy between BA.2 and BA.1 is present. Given that getting the booster shot can induce strong antibody production against Omicron7, it’s likely that the boosters will work well against BA.2 infection too.
“Deltacron” – the variant that wasn’t
Now, the rapid emergence of BA.2 comes on the heels of Leondios Kostrikis, a professor of biology at the University of Cyprus, reporting the detection of a new variant infecting 25 people a few weeks ago8. The variant, dubbed as “Deltacron” by social media, apparently had genomes comprised of Omicron-derived genetic material in a Delta-variant genome background. Before continuing, one must be careful with the name “Deltacron” here. Dr. Kostrikis never coined the name “Deltacron”, did not claim the virus as a hybrid between Delta and Omicron, nor was genetic recombination posed. Dr. Kostrikis only claimed that genomes comprised of genetic material from Omicron and Delta were present among patients infected by both Omicron and Delta9. Nevertheless, little more has been made of this supposed variant since Dr. Kostrikis’s report. In fact, “Deltacron” has fallen out of discourse in favour of the emerging Omicron BA.2 infections taking place worldwide. What happened to “Deltacron” then? It seems that the scientific community concluded that this supposed variant was likely the result of sequencing contamination10.
But how did the sequence contamination occur?
To better understand how sequence-based contamination can arise when sequencing SARS-Cov-2 genomes, a basic understanding of how the samples are prepared for sequencing is needed. PCR amplification represents the first step for preparing SARS-Cov-2 genomes after obtaining the genetic material from clinical samples (eg. nasal swabs and saliva) (Figure 1). In a typical PCR assay of SARS-Cov-2, the DNA generated through reverse transcription of SARS-Cov-2 RNA fragments (I will call them cDNA from now on) is amplified using small stretches of DNA – called primers – that amplify the SARS-Cov-2 cDNA. When using one pair of primers, only one region of the SARS-Cov-2 genome is amplified. A few single regions like these being amplified would help identify SARS-Cov-2 when searching for positive COVID-19 cases. However, when trying to sequence an entire genome, hundreds more primers are needed to sufficiently amplify every stretch of cDNA belonging to SARS-Cov-2 after reverse transcription. That’s because the ability to obtain enough SARS-Cov-2 RNA and cDNA, in turn, relies on being able to amplify all cDNA belonging to SARS-Cov-2 in the clinical samples.
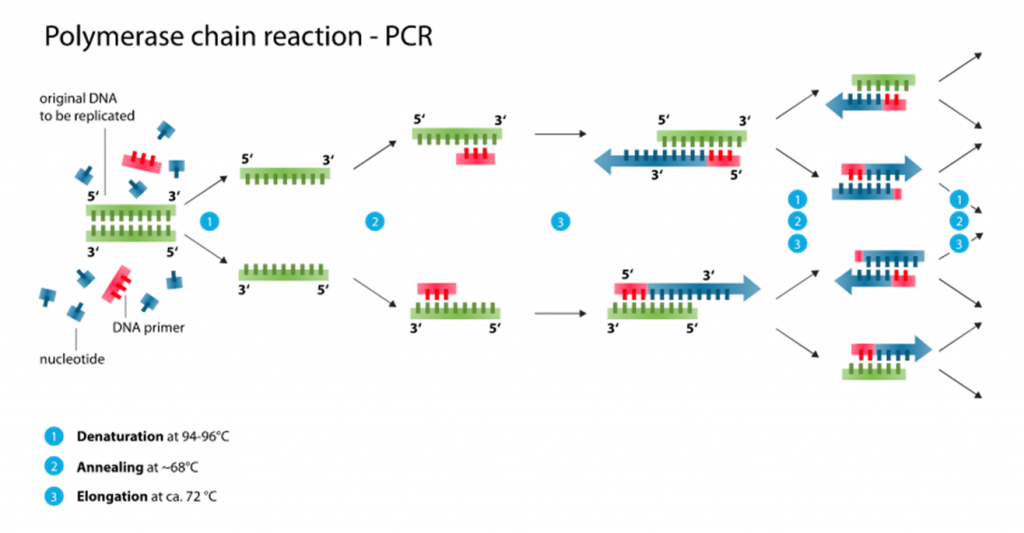
Now let’s move on to PCR amplification for SARS-Cov-2 genome sequencing (Figure 2). Here, hundreds of primers targeting different fragments of the SARS-Cov-2 genome are mixed in samples for PCR amplification. Many of the primers being used for amplification are only used to sequence SARS-Cov-2 as a whole, not a specific variant. At the time Dr. Kostrikis reported his new variant, Omicron-specific primers for sequence amplification were just being released. With hundreds of primers to use at once, the primers targeting Delta and the primers targeting Omicron may have been accidentally mixed before doing the PCR to begin the sequencing preparations. In particular, it’s possible that having Omicron-specific primers amplifying the spike protein would mix with primers targeting Delta for the rest of the genome. This is especially possible since Delta does not have the deletion at amino acids 69 and 70 of the spike protein that Omicron has. By the time the PCR and sequencing are done, a Delta-based genome with spike protein mutations observed in Omicron would end up getting sequenced. That was exactly what happened when scientists analyzed the genomes Dr. Kostrikis uploaded of his new variant. On top of that, clinical samples often have very low quantities of RNA for sequencing; these amounts can be as low as 1 nanogram (that’s 0.000000001 grams)! It would therefore also be possible to see a Delta genome contaminated with Omicron RNA if clinical samples from Omicron-infected and Delta-infected patients were accidentally mixed when processing multiple samples at once. While that possibility may be less likely, these two possibilities provide a plausible explanation for “Deltacron” not to have been detected anywhere else aside from the lone Cypriot lab.
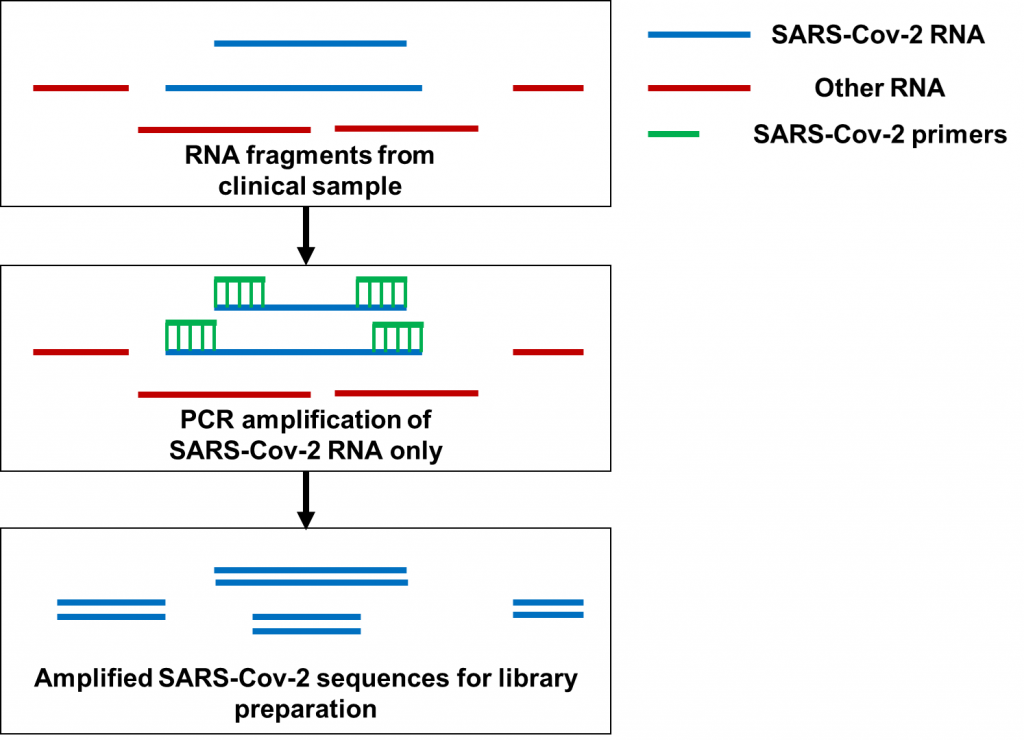
The contamination saga associated with “Deltacron” provides an important reminder to ensure proper adherence to sequencing preparation protocols. When preparing genetic material for sequencing, proper care has to be made to ensure that contaminants do not get sequenced. This includes using materials free of DNA and RNA contamination, making sure that samples are not mixed up, and ensuring that primer combinations to identify specific variants are not mixed. As a PhD candidate who worked on RNA sequencing for Pseudomonas aeruginosa, I worked with quantities as low as 10-100 ng of RNA and made every effort to make sure that RNA from other bacteria didn’t make its way into my sequencing preps. I had to be especially careful because our whole lab was filled with bacteria cultured from fecal matter and sputum! Imagine working with even lower amounts of RNA for sequencing, commonly encountered when working with respiratory specimens like nasal swabs.
References
- Aziz, Saba. January 25th, 2022. Global News. https://globalnews.ca/news/8538300/omicron-subvariant-ba-2-explained/
- Passafiume, Alessia. January 26th, 2022. The Toronto Star. https://www.thestar.com/news/canada/2022/01/26/health-officials-say-51-cases-of-omicron-ba-2-variant-detected-in-canada.html
- UK Health Security Agency. January 21st, 2022. UK Government. https://www.gov.uk/government/news/covid-19-variants-identified-in-the-uk
- Naphtali, Paul. January 11th, 2022, Microbe Musings https://microbemusings.ca/2022/01/11/a-picture-of-omicron-spread-in-south-africa/
- Clinical Conversion Staff. January 14th, 2021. ThermoFisher Scientific. https://www.thermofisher.com/blog/clinical-conversations/the-s-gene-advantage-taqpath-covid-19-tests-may-help-early-identification-of-b-1-1-7/
- Bernstein, Lenny. January 26th, 2022. The Washington Post. https://www.washingtonpost.com/health/2022/01/24/covid-omicron-ba2/
- Pajon, R., Doria-Rose, N.A., Shen, X., Schmidt, S.D., O’Dell, S., McDanal, C., Feng, W., Tong, J., Eaton, A., Maglinao, M., et al. (2022). SARS-CoV-2 Omicron Variant Neutralization after mRNA-1273 Booster Vaccination. New England Journal of Medicine 0, null.
- TOI Staff. January 9th, 2022. Times of Israel. https://www.timesofisrael.com/cyprus-identifies-combined-deltacron-covid-strain-but-says-unlikely-to-spread-far/
- Georgiou, Georgios. January 9th, 2022. Bloomberg. https://www.bloomberg.com/news/articles/2022-01-09/cypriot-scientist-says-covid-19-variant-deltacron-not-an-error
- Nature. January 21st, 2022. https://www.nature.com/articles/d41586-022-00149-9
