The Enigma Code and genome annotation
In World War II, Nazi Germany designed a cipher machine that encrypted all messages sent between military officials. It was only after years of hard work by the group at Bletchley Park, led by Alan Turing, that would finally crack the Enigma code. Key to the successful decoding of the Enigma Code was the weather transmissions that the British intercepted each day. In those transmissions, the same set of decoded words was daily sent at the end of each report that would help crack the entire code. You can learn about this effort in greater detail in the 2012 film “The Imitation Game” and this excellent two-part video series from Numberphile (Part 1 and Part 2)1.
Microbiologists specializing in computer-based skills face a similar decoding problem every day. Like Turing’s team, these microbiologists are tasked with cracking a code of their own. But the code that these scientists are cracking is not written in a spoken human language. Instead, they have to decode the language of genes. For microbiologists, decoding is even more challenging than cracking Enigma in two ways: 1) where the code starts and ends is not readily apparent, and 2) we still have not characterized the full set of proteins made in every bacterial species.
A protein to a human is not the same protein to a biochemist
To understand what it means for a gene to be encoded, we need to understand what proteins are. Firstly, all living beings (you, me, and microbes too) produce proteins. In biochemistry, proteins are not the pieces of meat we enjoy for a meal. Instead, a protein is like a molecular LEGO set (Figure 1). To make a LEGO toy, we’d have to put LEGO pieces together in a specific way. Amino acids are like LEGO pieces in that they come in different shapes and sizes. These amino acids also fit together in a specific way to make a chain that folds into a functional protein. LEGO toys are used for play or exhibition, but proteins can do all kinds of things that allow life to emerge.
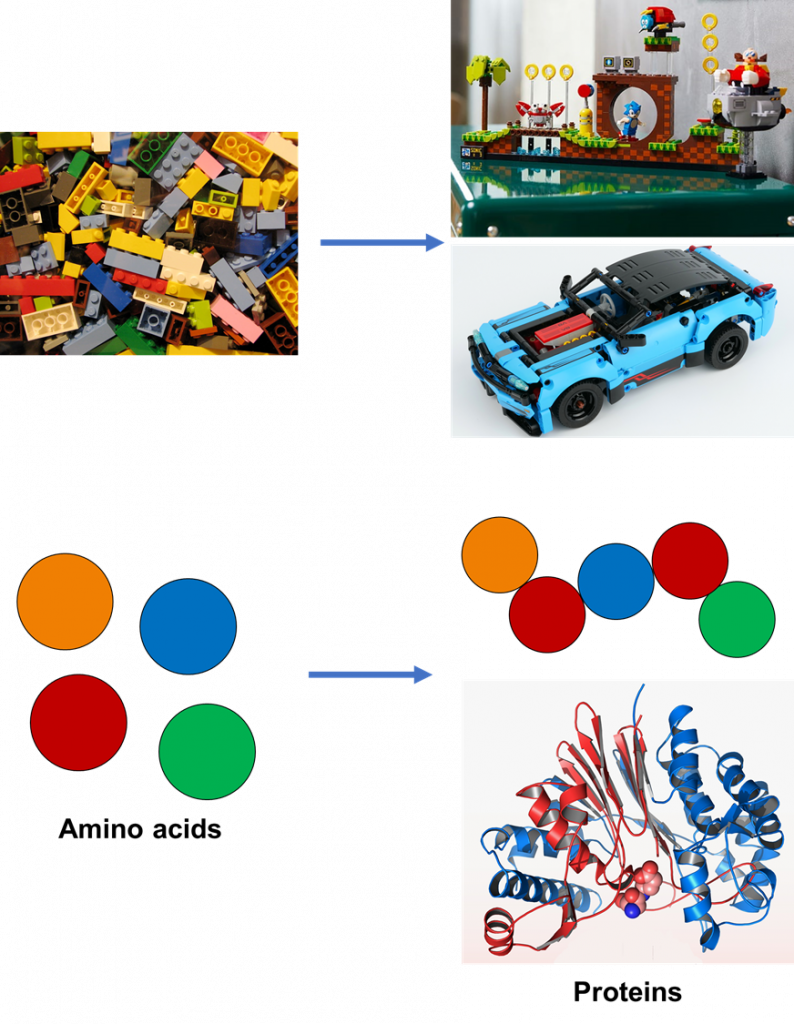
The genetic language for life
Like a LEGO set, proteins are constructed from a manual. These manuals are encoded in a set of genetic instructions called the genome. The manuals are written only with four letters: A, T, G, and C. To make the protein, the DNA manuals are first rewritten into RNA using the letters A, U (replaces the T in an RNA message), G, and C. Then, a biological translator, also known as ribosomes, takes the RNA manual and translates its instructions into a functional amino acid chain. The chain then folds to become a protein.
On its own, the DNA manual does not come with spaces. So how can the translators translate the message in the DNA (Figure 2)? The translators search for a stretch of three letters that starts the message: AUG. From the start of the message, the translator reads the DNA as three-letter words, translating each word into a single amino acid. The translating continues until the translator comes across one of three words: UAG, UGA, and UAA. At that point, the translator knows that the message has ended, and the translation stops.

The two components of genome annotation
The human genome can encode as many as 20,000 proteins. For their microscopic size, a microbial genome can encode as many as 6,000 proteins. Scientists know these numbers because they made painstaking efforts to decode their genetic messages. This decoding process is known as genome annotation. Genome annotation involves knowing where every genetic message starts or ends and which genes encode which proteins. These two components are called gene prediction and protein annotation respectively.
Gene prediction is only possible because we know that a genetic message typically starts with ATG and ends with a combination of TAG, TGA, and TAA. However, the microbe that I study has a genome over 6 million letters long. Furthermore, these messages can be read forwards and backwards. Searching through millions of letters for messages would not be possible for me if I searched through each letter manually. Such was also the case for Turing’s team tasked with deciphering thousands of encrypted German messages. In response, Turing’s team designed a large computer specifically for deciphering thousands of German messages. Similarly, scientists have developed programs to automate gene predictions. These programs search for ATGs and generate as many strings of three-letter words as possible to decrypt the encoded protein messages.
Once the protein messages have been deciphered, the next step is to know which sets the protein belongs to. Every purchased LEGO set has a specific code and theme within LEGO’s catalogues, like LEGO Harry Potter or LEGO Star Wars. Like the LEGO sets, proteins have specific names. Proteins can also be clumped into families depending on their traits. These characteristics are archived in protein databases and are used to determine a protein’s name based on the amino acid message. This process is called protein annotation and makes up the other half of the genome annotation.
The ways that genome annotation is harder than Enigma
Now that I explained what proteins are and where they come from, we can come back to comparing genome annotations with the Enigma Code. Every message has a beginning and an end. Such was the case for every Enigma-encrypted transmission and genes encoding proteins. However, messages encoded in a genome do not have a clear heading or footer that points to the start and end of the messages. While Turing knew that every intercepted transmission was a real message with a clear start and end, gene prediction tools do not have that luxury. With the spike protein example in Figure 2, the protein message could begin in at least 4 locations. However, the true encoded message begins where the green line is located and provides the instructions for making the SARS-Cov-2 spike protein. If the genetic message began at any of the red lines, the resulting protein message would be gibberish. Gene prediction tools must therefore learn from a training set of genetic messages confirmed to be readable. The tools would then predict which sets of protein messages are true and which ones are gibberish.
Most human language is designed so that people can understand each other when letters form words. But what if one wasn’t fluent in the language to be translated? For instance, even if Turing’s group successfully decrypted the code into German, not knowing the German words would have left them without a way to know what the messages mean in English. Microbiologists regularly encounter this kind of problem whenever they come across a poorly characterized protein. From a language standpoint, microbiologists know that there’s probably a protein sequence, but have little idea of what the protein is. This problem is especially prevalent for many microbes, where as little as 14% of the protein messages have been accurately deciphered and profiled2. Although microbiologists are hard at work to characterize these proteins and study the protein language, much more needs to be done to fully understand the protein language that microbes express.
Why is genome annotation important?
Understanding the genetic and protein language that microbial life speaks is essential for understanding how microbes behave in all kinds of growth conditions. Without a robust genome annotation, we would not be able to learn how SARS-Cov-2 infects humans so quickly, treat COVID-19, or even make the vaccine in the first place. Robust genome annotation is vital for knowing what sets of proteins to target when developing antibiotics to fight infectious disease. Finally, we would not be able to determine the ways through which probiotics can help treat intestinal diseases like IBD or Crohn’s disease. The many ways through which genome annotation drives scientific discovery in microbiology make genome annotation a worthy venture, no matter how difficult it is.
References
